Brain Tumor Classifier
Abstract
This project focuses on developing a deep learning model to classify brain tumors using MRI scans. By leveraging transfer learning techniques with pre-trained convolutional neural networks, we achieved a high diagnostic accuracy of up to 99%. The model differentiates between glioma, meningioma, pituitary tumors, and healthy brain tissue. Our results highlight the potential of AI in enhancing early detection and diagnosis in neuro-oncology, providing valuable support for clinical decision-making. The project also emphasizes the importance of dataset curation and model interpretability, ensuring the reliability and trustworthiness of AI-driven healthcare solutions. This project can assist neuro-oncologists in real-world scenarios. All project resources are available on GitHub and Kaggle for further exploration and collaboration.
Introduction
Distinguishing between healthy brain tissue and various types of tumors, such as glioma, meningioma, and pituitary tumors, is a critical challenge in neuro-oncology. Early and accurate classification of MRI scans provides essential information for treatment planning, reducing risk, and potentially improving patient outcomes. In this project, we leveraged deep learning, specifically transfer learning techniques, to build a classification system capable of differentiating four categories in MRI brain images: glioma, meningioma, pituitary tumor, and no tumor. By utilizing pre-trained convolutional neural networks (CNNs) and fine-tuning them on our curated dataset, we aimed to achieve a high level of diagnostic accuracy (99%) that could support clinical decision-making.
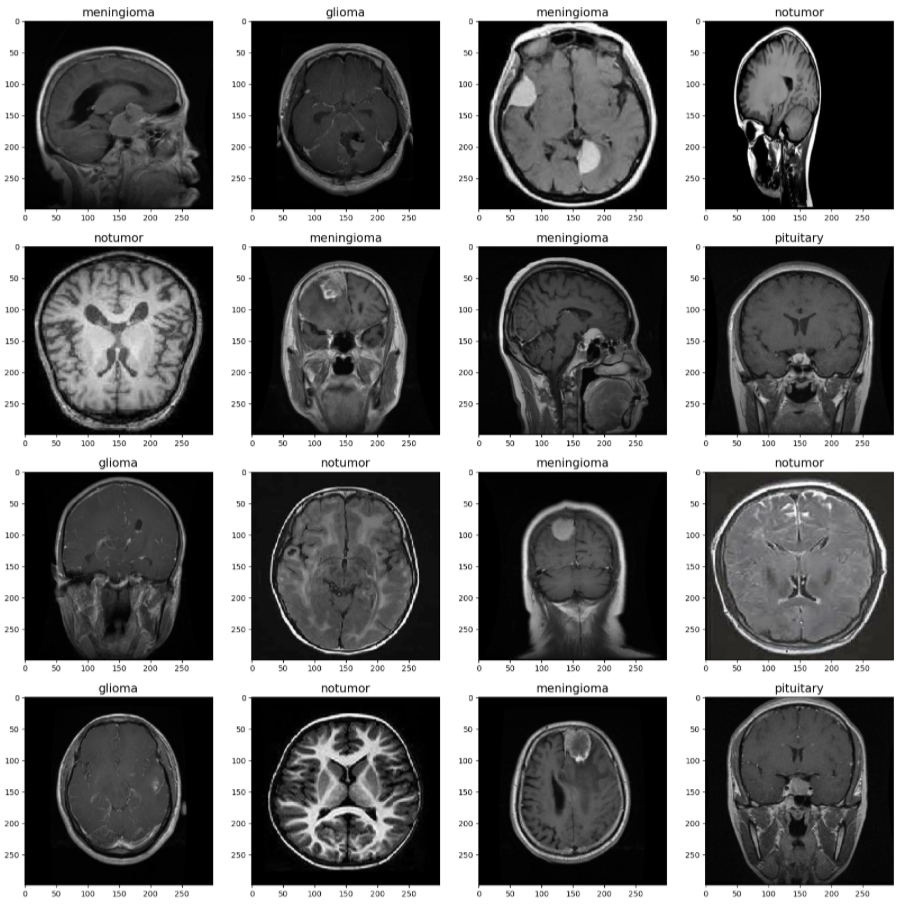
Background
Common types of brain tumors include: Gliomas arise from glial cells, which support neuron function. Meningiomas form in the meninges, the protective layers of the brain. Pituitary tumors develop in the pituitary gland, which regulates hormones. The absence of pathological growths indicates a healthy brain with no tumor. Magnetic Resonance Imaging (MRI) is a non-invasive imaging modality commonly used to diagnose brain abnormalities. Our project focuses on leveraging Machine Learning (ML) and Deep Learning techniques—specifically transfer learning with pre-trained convolutional neural networks—to classify MRI images into four categories: glioma, meningioma, pituitary tumor, or no tumor. The goal of this project is to develop a robust ML model capable of accurately classifying brain MRI images into these four classes. The motivation is to help clinicians with early tumor detection, supporting more effective and timely treatments and improving patient quality of life.
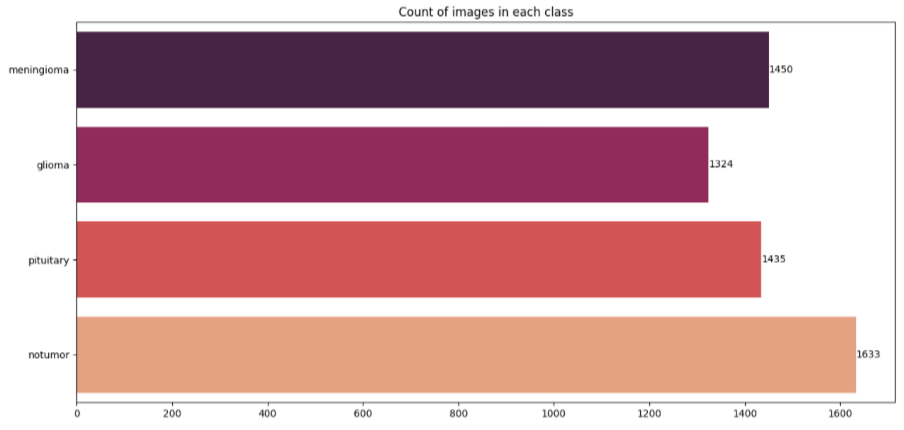
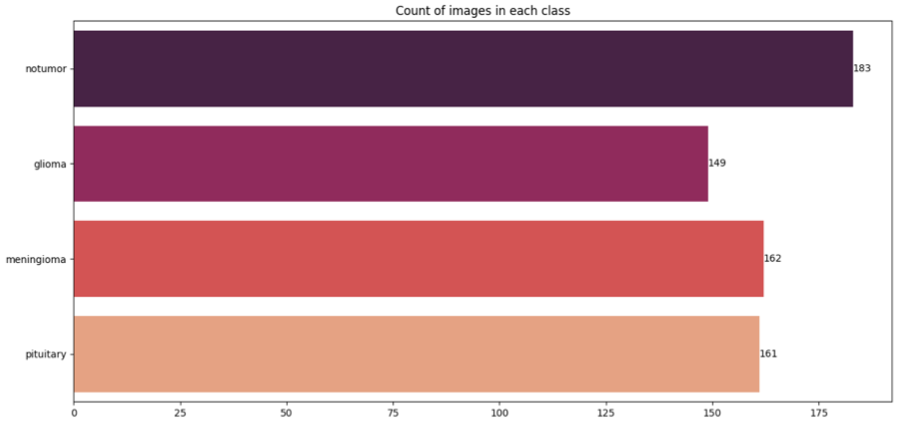
Data and Preprocessing
Our dataset was created by combining and curating images from multiple publicly available sources on brain tumor:
- Figshare Brain Tumor Dataset
- Kaggle: Brain Tumor Classification MRI (Sartaj Bhuvaji)
- Kaggle: Brain Tumor Detection (Ahmed Hamada)
Modeling
The core of our modeling strategy involved transfer learning with well-known architectures trained
initially on
ImageNet, a large-scale natural image dataset. We chose to compare the performance of two
architectures: VGG16 and
Xception. Both models are widely respected for their robustness and representational power. By first
“freezing” all
or
most convolutional layers, we preserved the general feature extraction capabilities learned from
millions of natural
images, then fine-tuned only the top layers to specialize the models for MRI-based tumor
classification.
In practice, we worked primarily in a Python environment leveraging the TensorFlow and Keras
frameworks. Our
notebooks
detail every step of this process. For instance, we implemented data generators to feed images to
the network. Using
the
ImageDataGenerator class, we employed rescaling and optional brightness adjustments. During
training, we employed a
batch size of 32 and ran for 10 epochs initially. The models utilized the Adamax optimizer at a
learning rate of
0.001,
chosen after preliminary testing indicated stable convergence. We monitored training progress via
accuracy, loss,
precision, and recall metrics, all logged at each epoch. Validation performance guided early
stopping criteria and
hyperparameter refinements.
Metrics for Xception model:
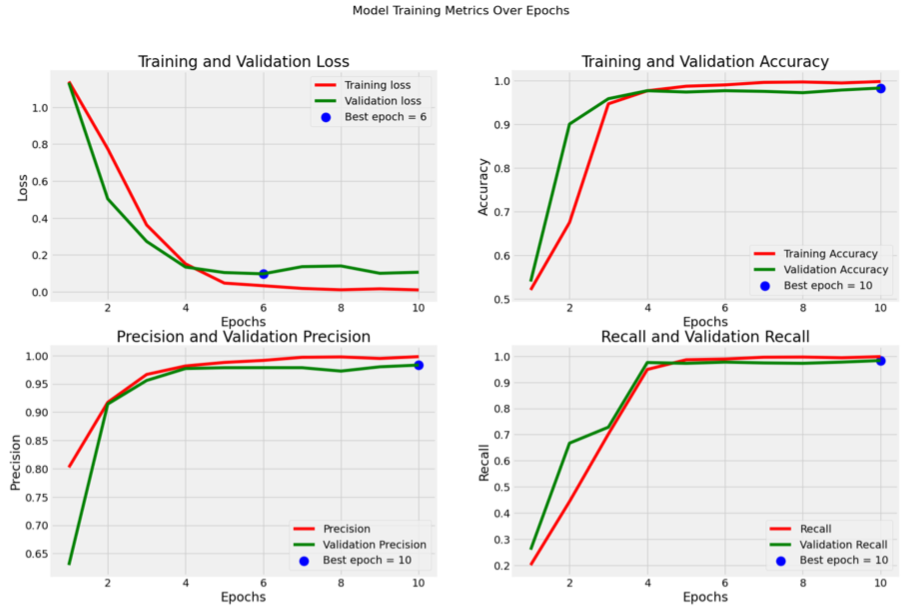
Results
The final results indicated that transfer learning is an effective strategy for MRI-based brain
tumor
classification.
The Xception model, in particular, demonstrated a remarkable aptitude for extracting and
internalizing relevant
features, achieving over 99% accuracy on the test set. These findings suggest that, given a
sufficiently large and
clean
dataset, pre-trained CNNs can be adapted to medical imaging tasks with minimal architectural changes
and relatively
short training times.
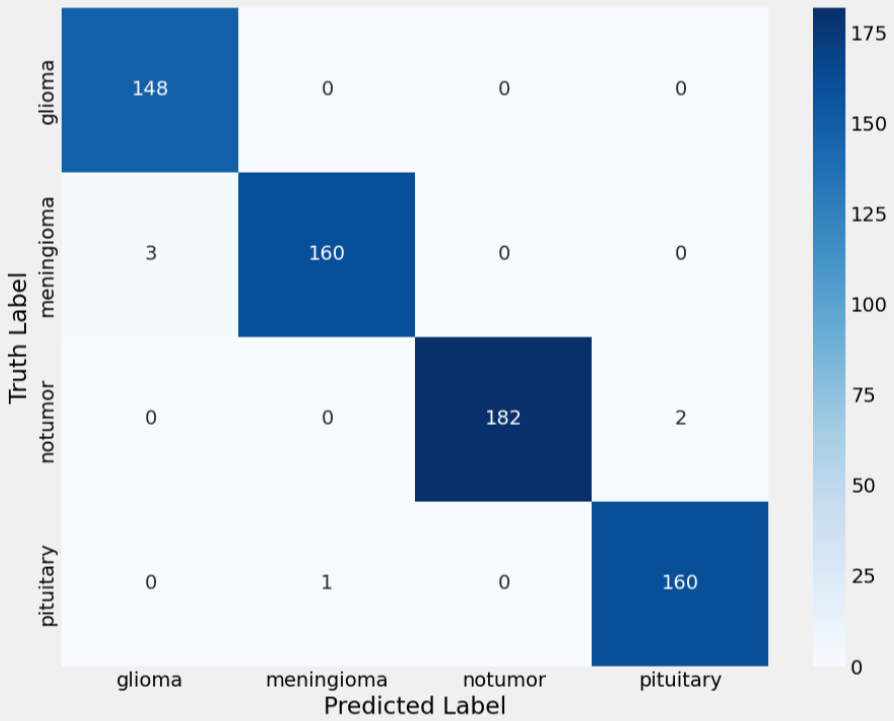
Challenges and Future Work
This project presented several challenges. Curating a large, high-quality dataset was time-consuming but vital for model reliability. The reliance on GPU resources was essential, as the complexity and size of these models demanded significant computational power. Training took about 45 minutes on an H100 GPU. Iterative tuning of hyperparameters—careful adjustments to the learning rate, batch size, and number of unfrozen layers—was necessary to achieve optimal results. Despite these hurdles, the outcome was successful and instructive, showing that perseverance and systematic experimentation can yield strong performance in medical imaging tasks. Moving forward, there are several promising avenues for extending this work. Testing the model on external datasets would establish its robustness and clinical relevance. Implementing newer architectures like EfficientNet or Vision Transformers might further improve accuracy and generalization. Additionally, incorporating meta-data, such as patient age or clinical histories, could enhance diagnostic power. Above all, exploring interpretability methods would help bridge the gap between cutting-edge machine learning and actionable clinical insights. The aim is also to collaborate with other ML engineers to explore new ways to make the model more efficient, robust, and faster, and to enhance the dataset so it is suitable for segmentation tasks.
Project Resources